
Few subjects in archaeology capture the imagination like Egyptian mummies do. These beautifully preserved bodies, particularly those of kings and queens, offer a tantalising window into the ancient past. And now, with state-of-the-art scientific techniques such as X-ray scanning and genetic analysis, we are finally getting definitive answers to questions relating to health, family relationships, and causes of death.
Or are we? Despite a string of triumphant headlines, the scientific analysis of Egyptian mummies remains incredibly difficult. The truth is that the latest studies are - so far - only making our understanding murkier.
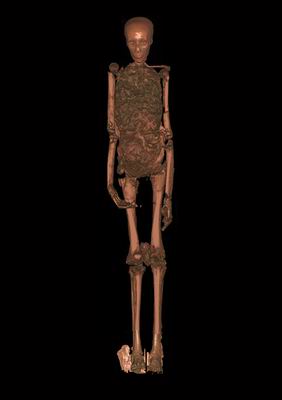
I've just written a feature for New Scientist (What killed Tutankhamun - sub required) about efforts to determine the cause of death of Tutankhamun, who died after a short reign in around 1333 BC. After decades of wild speculation about everything from birth defects to murder, 3D X-ray scans published in 2005 (see second pic, below) identified a broken leg (Annales du Service des Antiquités de L'Égypte, vol 81, p 159). Then a DNA analysis published in 2010 concluded that the pharaoh was an inbred genetic weakling, and infected with malaria (Journal of the American Medical Association, vol 303, p 638).
Both studies gained huge press coverage, including glitzy TV documentaries, and the findings have entered popular wisdom about Tutankhamun. But there is a large dollop of uncertainty associated with these results. For example, the broken leg could have been fractured after Tut's death, by the royal embalmers, or after the mummy was unwrapped in 1925.
But it is the DNA findings that, behind the scenes, are causing the most debate, with some experts concerned that all the researchers have detected is modern contamination. I've written about this in a news story for New Scientist today (free to read) but as this is such a high-profile study I wanted to use this post to look at the arguments in a bit more depth, so you can make your own mind up!
This was a huge study of 11 royal mummies dating from around 1300 BC - the first DNA analysis of ancient Egyptian royalty - and it was carried out at great expense. The mummy samples aren't allowed to leave Egypt, so a state-of-the-art DNA lab was built at the Egyptian Museum in Cairo, funded by the Discovery Channel, which filmed the proceedings. The study was led by Zahi Hawass, the exuberant head of Egypt's Supreme Council of Antiquities, and carried out by an Egyptian team, with the help of two international consultants, Albert Zink of the EURAC Institute for Mummies and the Iceman in Bolzano, Italy, and Carsten Pusch of the University of Tübingen, Germany - both experienced mummy researchers.
The results were published with great fanfare in February 2010, alongside the Discovery documentary. The findings were suitably dramatic. As well as detecting DNA from the malaria parasite in four of the mummies, the researchers produced a family tree. Among other things, they identified Tut's father as the heretic pharaoh Akhenaten, concluded that Tut's parents were brother and sister, and found that two mummified foetuses found in Tut's tomb were probably his stillborn children.
It was an unprecedented wealth of information about Tutankhamun and his family, and the authors described their work as heralding a new scientific discipline, of "molecular Egyptology".
What hasn't made the news until now is that for many in the ancient DNA community, this study has triggered not excitement but scepticism and frustration.
 To start with, some researchers are convinced that it is theoretically impossible for DNA to survive so long in such a hot environment. The warmer the conditions, the faster DNA degrades. So although DNA has been recovered from frozen mammoth or Neanderthal remains that are tens of thousands of years old, it is not expected to last as long in a hot tomb (probably around 30 ˚C). Ian Barnes, a molecular palaeobiologist at Royal Holloway, University of London, and his colleagues have calculated that in such conditions it would only last a few centuries at most, and certainly not for 3,000 years (American Journal of Physical Anthropology vol 128, p 110).
To start with, some researchers are convinced that it is theoretically impossible for DNA to survive so long in such a hot environment. The warmer the conditions, the faster DNA degrades. So although DNA has been recovered from frozen mammoth or Neanderthal remains that are tens of thousands of years old, it is not expected to last as long in a hot tomb (probably around 30 ˚C). Ian Barnes, a molecular palaeobiologist at Royal Holloway, University of London, and his colleagues have calculated that in such conditions it would only last a few centuries at most, and certainly not for 3,000 years (American Journal of Physical Anthropology vol 128, p 110).
Because of this, Barnes and others say that they would expect the resulting paper to provide extensive information about the methods used, and the efforts taken to rule out modern contamination. They would also like to see the raw data, to judge the validity of the results for themselves.
Unfortunately, perhaps because the paper was published in a medical journal rather than one that specialises in ancient DNA, such information was not included. "There's a lack of detail which I find quite puzzling," says Eline Lorenzen of the Centre for GeoGenetics at the Natural History Museum in Copenhagen, Denmark, who wrote to JAMA in June, expressing caution over the results (vol 303, p 2471). "I would be surprised if any of the paper's reviewers were within the field of ancient DNA," she says. "Many of us in the DNA community are really surprised that this has been published."
Critics are especially concerned by the method that Hawass's team used to analyse the mummy DNA. The DNA in ancient samples is generally degraded, present in very small amounts, and contaminated with modern DNA. This is a particular problem for human samples, where you have to work out whether the DNA you have belongs to the original individual or to other people who have come into contact with the body in modern times.
Ancient DNA researchers usually start by trying to amplify and sequence mitochondrial DNA (mtDNA). There are thousands of copies of this in every cell, so it is much easier to work with than genomic DNA, of which there are only two copies in every cell.
Mitochondria are passed down the maternal line, so they provide information about family relationships. And sequencing the DNA gives a sense of the quality of the sample - for example it will be obvious whether you have just one sequence, or a mix of contaminating DNA from different individuals.
But the JAMA paper doesn't mention mtDNA. Instead, the researchers used genetic fingerprinting to construct their family tree. You might think that genetic fingerprinting, famous for its use in criminal investigations, should give black-and-white results. But it can actually be very subjective, particularly for poor quality samples (for example see Linda Geddes' excellent investigation).
Genetic fingerprinting involves testing variable regions of the genome called microsatellites. These are made up of short sequence repeats, the exact number of which differs between individuals and is inherited from parent to child. Each microsatellite region is amplified using a technique called polymerase chain reaction (PCR), then researchers estimate from the size of the product how many repeats it must contain. By comparing individuals over a number of such regions, it's possible to work out whether or not they are related.
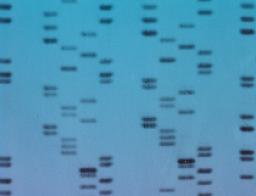 The problem is that PCR amplifies modern contamination as well as ancient DNA, and simply checking the size of the products offers no way to distinguish between the two. "With microsatellites, it is very hard to establish if there is contamination," says Barnes. "They don't give you the sequence, it is just a coloured band on a computer screen."
The problem is that PCR amplifies modern contamination as well as ancient DNA, and simply checking the size of the products offers no way to distinguish between the two. "With microsatellites, it is very hard to establish if there is contamination," says Barnes. "They don't give you the sequence, it is just a coloured band on a computer screen."
To make matters worse, the PCR can slip on the sequence repeats, producing "stutter bands" of different sizes. For modern, good-quality samples, researchers can usually work out which are the stutter bands and which are the real ones. But for poor quality samples, especially when mixed with contamination, where there may or may not even be any original DNA present, teasing out which bands are real and which aren't is fraught with difficulty. "Very few ancient DNA studies have used microsatellites because they are so prone to genotyping errors," says Lorenzen.
She and Barnes are both keen to see the raw data so they can judge its quality for themselves. But the JAMA paper simply gives the genotypes that were eventually derived for each mummy.
As well as the raw data, Lorenzen wants more information about the precautions taken to guard against contamination in the first place: "How were the DNA samples taken? What measures were taken to ensure no contamination from the individuals doing the sampling? In modern-day Egypt, what are the frequencies of these alleles? If anything else was found in the graves, have they genotyped that? As far as I can tell they did not carry out any pre-treatment precautions. There is a complete lack of information about how this was done. It rings alarm bells."
Barnes agrees. "We know that the material has been handled in the past by quite a lot of people, and we know that DNA preservation is theoretically poor in this material. You have to assume that the material is already contaminated and then explain why the results tell you that you haven't got contamination."
 So what do the researchers have to say about all this? Hawass (pictured left, with Tut's mummy) sees no cause for concern. He told me that he is confident in the conclusions reached, and describes the recent studies as "the final word" on Tutankhamun. "We announced everything that we discovered only after the findings had been reviewed by scholars connected with respected scientific journals."
So what do the researchers have to say about all this? Hawass (pictured left, with Tut's mummy) sees no cause for concern. He told me that he is confident in the conclusions reached, and describes the recent studies as "the final word" on Tutankhamun. "We announced everything that we discovered only after the findings had been reviewed by scholars connected with respected scientific journals."
For a more in-depth response to the criticisms, I went to Pusch and Zink, who designed and oversaw the JAMA study. On how the DNA could have survived, both agree that DNA from a body simply buried in the sand would probably not survive from ancient Egyptian times. But they argue that in the royal mummies, the embalming process used by the Egyptians must have acted to preserve the DNA. This included drying out the body with a naturally-occurring mixture of salts called natron.
"The Egyptians really knew how to preserve a body," says Zink. "The worst thing for DNA is humidity. They got rid of the humidity very fast, and then protected the body from the re-entry of humidity, by covering it with oil, wax and bandages."
Pusch also believes that the embalming materials may themselves be acting to protect the DNA. "Nobody has thought about the components of the resin," he says. He says he is currently working with chemists at the University of Tübingen to investigate what different substances are present, and hopes to publish on this in mid-2011.
Regarding contamination, Pusch says that the team took their samples from deep inside the mummies' bones, where modern contamination could not have reached. (It's hard to see how this would have worked on the thin, fragile skeleton of Tutankhamun though, not to mention the tiny bones of the mummified foetuses.) They also genotyped the eight lab staff who carried out the genetic analysis to make sure that their DNA wasn't being picked up, and they checked every result in two independent labs. But as for Lorenzen's idea of using non-human remains as a negative control, "that would be a little bit stupid", Pusch says.
Pusch and Zink admit that their data, as to be expected for such difficult samples, was not always 100% clear. So they tested every microsatellite several times in samples from different locations on each mummy, and used a "majority rule" to decide on the result. "If we do the experiment 30 times on one mummy, we might get 18 the same," says Pusch. Then they used a computer programme to work out the most probable family tree for the resulting genotypes.
Zink says that he would be happy to discuss the results with other researchers but that he would be reluctant to share raw data, because the need to use the majority rule means "there could be a lot of arguing".
What about mtDNA? In fact, the researchers say that they have isolated mtDNA, but chose not to include it in the JAMA paper as they are still working on it. "We face problems getting clear results," says Zink. MtDNA sequences would give researchers their first insight into the genetic origins of the pharaohs - a potentially explosive issue - so "we want to be 100% sure", says Zink. He and Pusch are planning a paper on this, along with an analysis of the male mummies' Y chromosomes - later this year. The JAMA paper was "just the opening ceremony", says Pusch.
Overall, Pusch says he is disappointed by the criticism that the paper has received. "I don't understand people's harshness," he says. "These people have never worked on royal mummies. Give us a little bit more time, to show people that more is to come."
But Lorenzen is not swayed. "There has been so much coverage of this, but no mention of the criticisms," she says. "It is very frustrating."
So, is this one of the most exciting developments in Egyptology, and will 2011 bring our first insight into the genetic origins of the pharaohs? Or is this a case of researchers under huge pressure for results seeing what they want to see? Pusch and Zink seem to me to be talented and tenacious researchers working in very difficult circumstances. But they need to provide the methodological details and raw data that will convince other experts of the validity of their work. When publishing such important results you cannot expect to be given the benefit of the doubt.